Gene adaptation across scales
Current methods to detect adaptation focus either on the short time-scale (population genetics, e.g. human populations), or on the long time scale (phylogenetics, e.g. across mammals). It is still unclear whether the signatures of adaptation are congruent.
Using population-genetic formalism within phylogenetic methods (see notes on Mutation-Selection models), we predict the rate of evolution for genes and sites, called ω₀, accounting for purifying selection on amino-acids.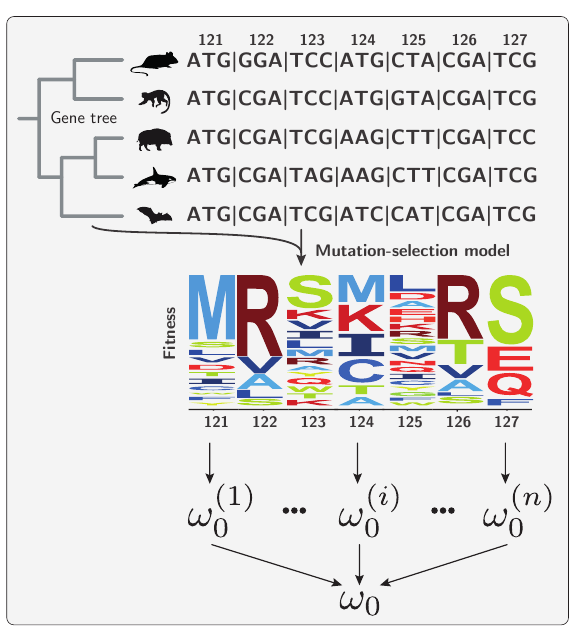
Formally and mathematically, ω₀ is the rate of evolution at which the gene is in mutation-selection equilibrium.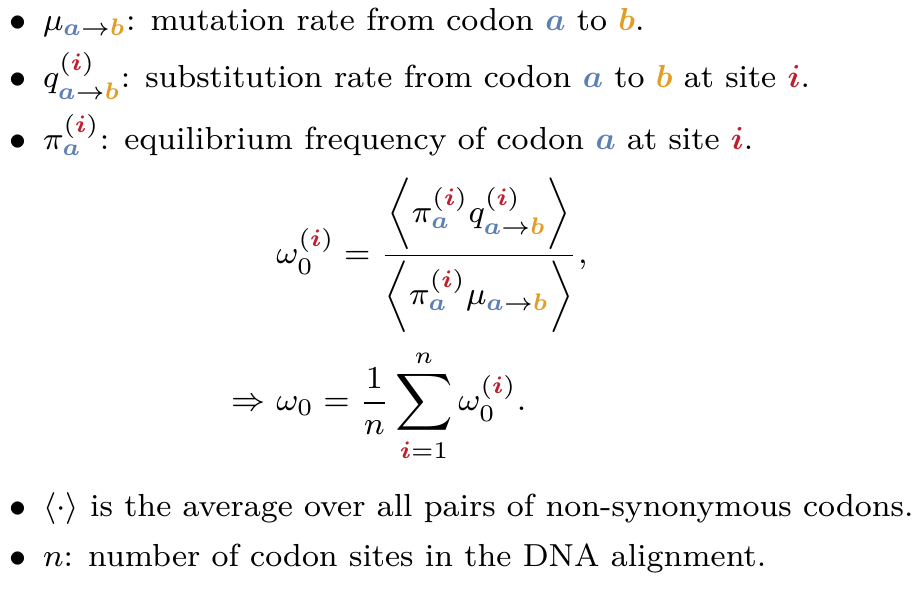
Low ω₀ imply heavy constraints on gene evolution. Genes for which the estimated rate of evolution (ω or dN/dS) is higher than ω₀ are evolving faster than expected, thus considered under adaptation.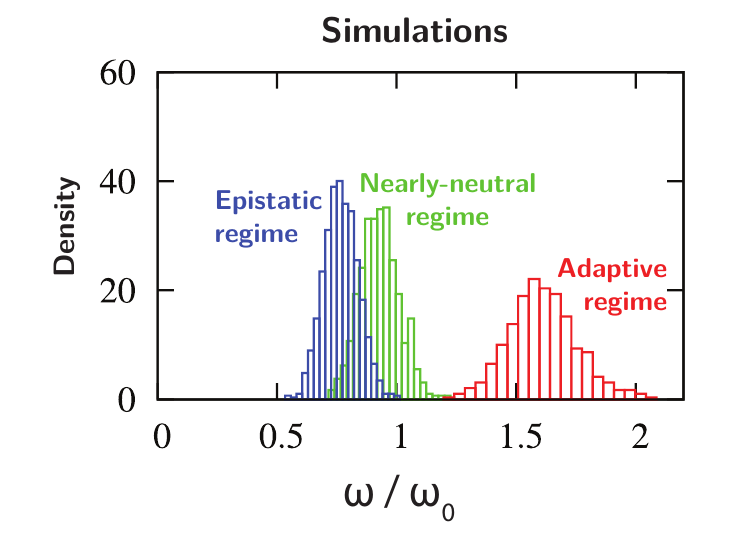
Note: using ω>ω₀ (gene is faster than predicted) instead of ω>1 (gene is faster than the neutral case) is statistically more powerful since ω₀<1.
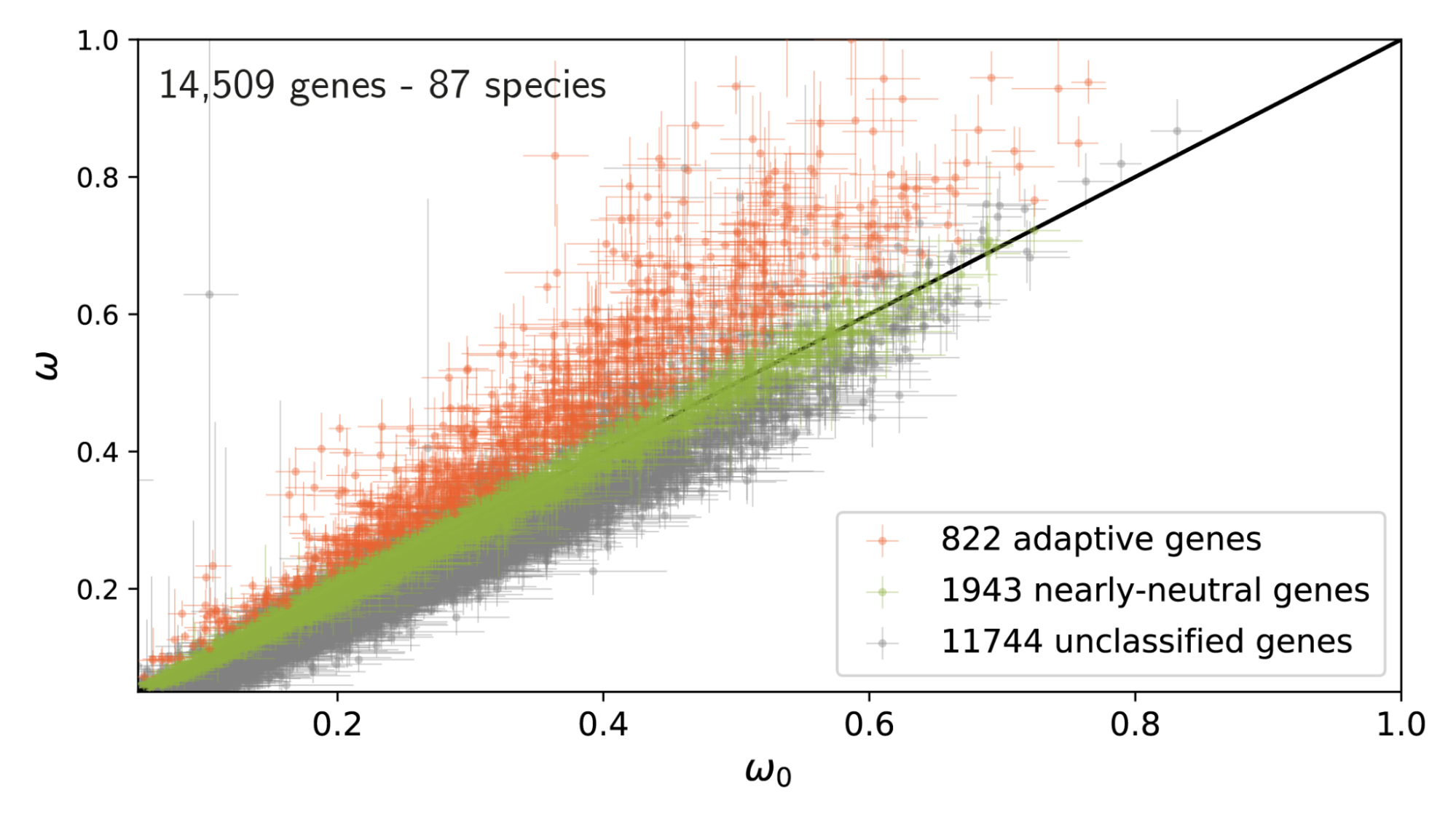
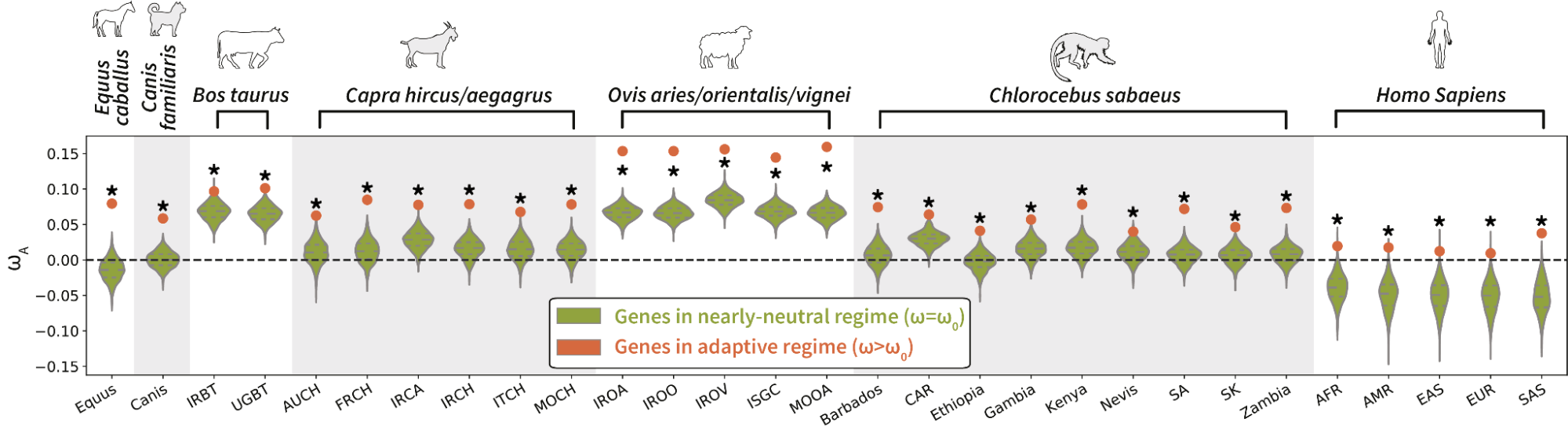
For genes under adaptation at the phylogenetic scale (red), we quantify the rate of adaptation at the population-genetic scale. This is compared to genes under a nearly-neutral regime (green).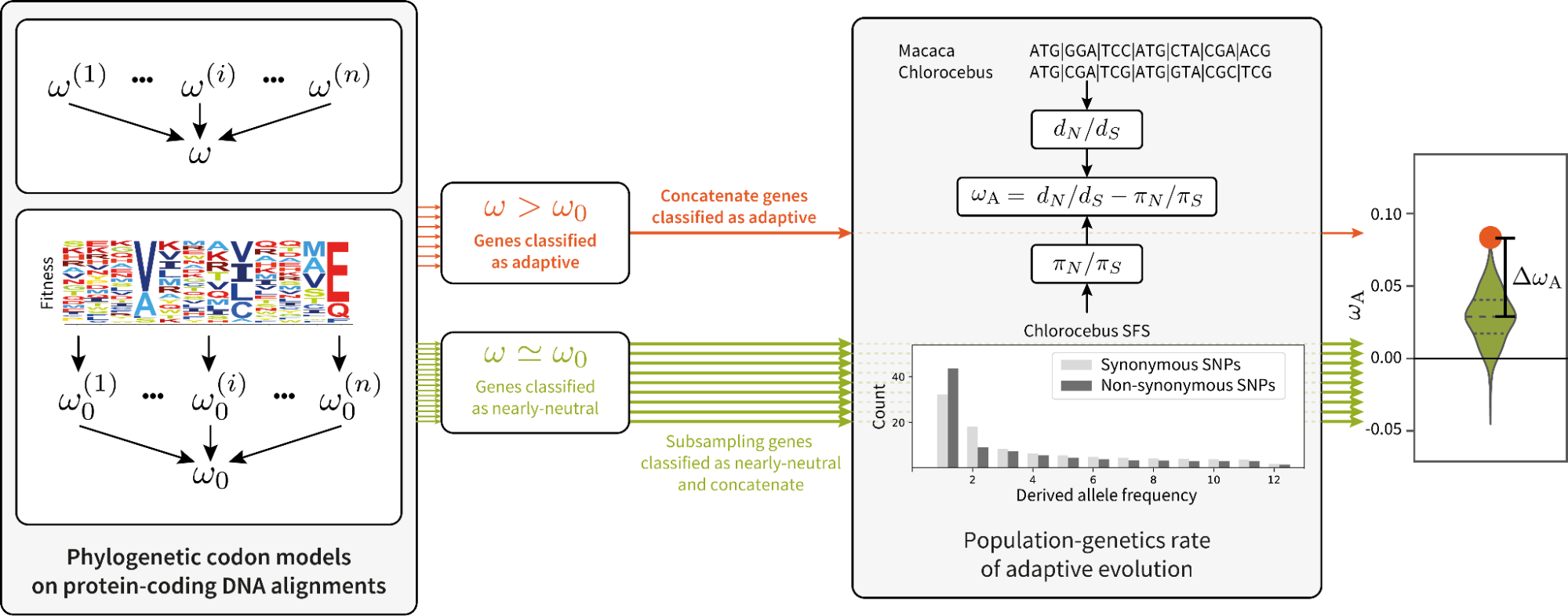
Across populations, the group of genes detected with a high rate of adaptation in the phylogeny-based method also displays a high rate of adaptation in the population-based method.